8. DFG-supported project - Role of DNA methylation for gene expression regulation in cyanobacteria
(HA 2002/17-1, joint project together with Prof. Wolfgang R. Hess, University Freiburg, project period 2015-2020)
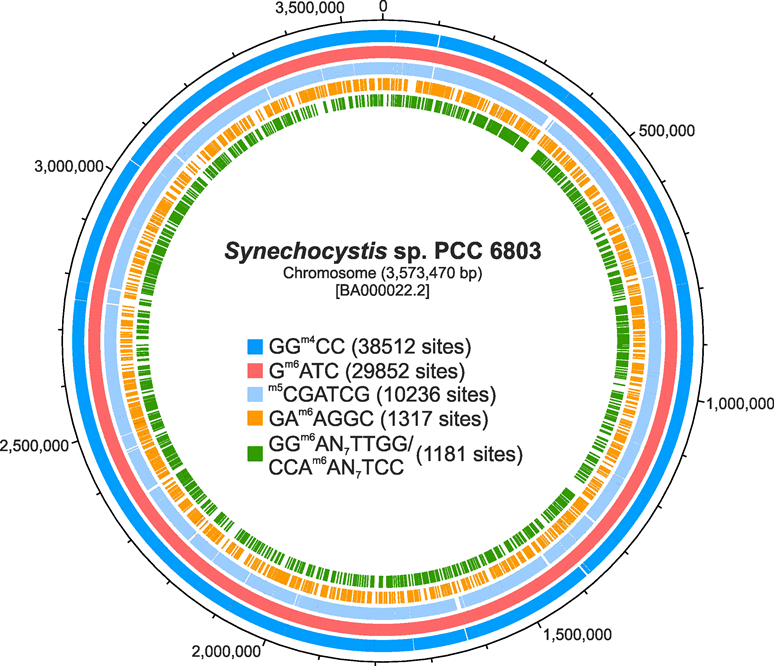
DNA methylation is currently receiving much attention as a key process for epigenetic modulation of gene expression among eukaryotes. In bacteria, DNA methylation is known as an important element of restriction-modification systems and of processes related to DNA repair, to the timing of DNA replication, or to chromosome partitioning. Various reports have discussed a possible regulatory function; however, there are no comprehensive studies on the impact of DNA methylation on gene expression among prokaryotes. The model cyanobacterium Synechocystis sp. PCC 6803 harbours genes for three orphan methyltransferases. Our own preliminary investigation of mutants defective in these DNA-specific methyltransferases MSsp6803I, MSsp6803II and MSsp6803III, encoded by the genes slr0214, slr1803 and sll0729, gave evidence that DNA methylation indeed plays an important role for gene expression regulation in this bacterium. In the proposed project we aim at investigating the mechanism underlying this regulation, the specificity and its integration into the response to changing environments. For this purpose, we plan to generate genes and promoters devoid or containing these methylation sites, to follow the dynamics of genome-wide methylation in response to different external factors, and to investigate these enzymes in vitro.






