1. DFG-supported project in the research consortium FOR2816 - SCyCode
The Autotrophy-Heterotrophy Switch in Cyanobacteria: Coherent Decision-Making at Multiple Regulatory Layers
Subproject: Metabolic regulation of carbon allocation from the CBB cycle under different CO2 supply (HA2002/23-1, project period 2018-2021)
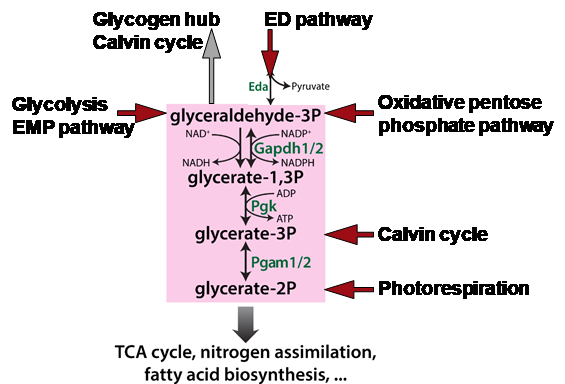
Cyanobacteria are photoautotrophic organisms, which assimilate CO2 during the day and use accumulated organic carbon by heterotrophic pathways during the night. Hence, anabolic and catabolic routes of primary carbon metabolism are both occurring in the non-compartimented cell, which need to be tightly regulated to avoid futile cycles. Cyanobacteria initially evolved in a CO2-rich environment, which became CO2 depleted and O2 enriched due to oxygenic photosynthesis. To adapt to this global change, cyanobacteria evolved an efficient carbon concentrating mechanism (CCM). Despite the cellular enrichment of inorganic carbon due to the CCM, cyanobacteria show a defined metabolic acclimation to low CO2 in the light. After shifts from high to low CO2 availability, Synechocystis increases the carbon flow out of the Calvin-Benson-Bassham (CBB) cycle into glycolytic routes and starts to consume glycogen reserves, which resembles the switch from autotrophic to heterotrophic modes after light/dark transitions. We assume that isoenzymes acting on branching points between CBB cycle and glycolysis such as phosphoglycerate mutases and glyceraldehyde-3-phosphate dehydrogenases as well as different enzymes involved in glycogen synthesis/breakdown are responsible for these alterations. The aim of this proposal is to identify regulatory mechanisms (e.g. protein phosphorylation and signal metabolites), which regulate anabolic and catabolic carbon fluxes at these branching points. Biochemical features of key enzymes and the impact of potential signal molecules such as the photorespiratory intermediate 2-phosphoglycolate will be studied in vitro using recombinant enzymes in cooperation with the Siebers team. To elucidate the impact of the different isoenzymes on cellular physiology, we plan to generate a defined set of mutants by deleting or over-expressing genes for specific isoenzymes and by replacing native enzymes with variants showing site-specific changes in biochemical target sequences, e.g. glycolytic enzymes with and without phosphorylation sites. These strains will be analyzed regarding alterations in the low CO2 acclimation and, together with the Kopka team, in the carbon flux by 13C-labelling. In cooperation with the Macek group the alterations in protein phosphorylation under different CO2 regimes will be studied and the involved protein kinases will be subsequently identified using our mutant library of all annotated protein kinase genes. Hence, we aim to elucidate mechanisms for the metabolic reprogramming of the cyanobacterial primary metabolism under fluctuating CO2 regimes via metabolic signals or via the modulation of key enzyme activities. The described topic will be analyzed in close collaboration of our network and will provide measures to manipulate the cyanobacterial metabolism for future biotechnological applications.